Course information for: BB485/585 Applied Bioinformatics
Computer Lab Projectes:Drosophila Genome data link: Drosphila genome version dm3. Download chromFa.tar.gz with the command at the terminal:
wget http://hgdownload.soe.ucsc.edu/goldenPath/dm3/bigZips/chromFa.tar.gz
Unzip the file with the command:
tar xvfz chromFa.tar.gz
Combine all the chromosome fasta files into one genome file:
cat chr*fa > dm3.fa
Build a blast database:
formatdb -t dm3 -i dm3.fa -p F -n dm3
Download the sequence infomation for human BRCA1 and create a fasta file for the sequence NCBI human BRCA1 link.
BLAST The sequence to the genome:
blastall -i brca1.fa -d dm3 -p blastn -o blast.dm3.out
Download the RefSeq mRNA annotations refMrna.fa.gz with the command at the terminal:
wget http://hgdownload.soe.ucsc.edu/goldenPath/dm3/bigZips/refMrna.fa.gz
gunzip the file with the command:
gunzip refMrna.fa.gz
Create a database for the RefSeq annotations:
formatdb -t refMrna -i refMrna.fa -p F -n refMrna
BLAST The sequence to the refMrna database:
blastall -i brca1.fa -d refMrna -p blastn -o blast.ref.out
Discussion questions: the difference between the two results? How do you explain the difference? What is chrUextra anyway?
Homework: How does this e-value compare to BLASTing to mouse through the NCBI website? Can you find a gene in human that has a significant hit to the E. coli genome?
download the perl script to print genome loci here: perl script
You can use this script along with the chromosome files you downloaded in Project 1 to pring genomic locations with the command:
perl printGenomicLocation.pl chr2R.fa 5873440 5874240
This particular genomic region is an eve stripe enhancer. You can find more eve stripe enhancers by going to the website/database: Redfly Go to this website, and enter "eve_stripe" for the Element Name/FBtp
build a fasta file containing all the eve stripe enhancers by appending these contenets of each genomic region to a fasta file. For example:
perl printGenomicLocation.pl chr2R.fa 5873440 5874240 > enhancers.fasta
perl printGenomicLocation.pl chr2R.fa 5865267 5865750 >> enhancers.fasta
...
and so on.
First create a fasta file with binding sites for twist. Go to the redfly database from Project 2. Enter "twi" into the Element name box. Click search, then in the search results select the TFBS tab.
Double click the entries with "twi" in the TF name column. In the new window that pops up within the webpage, click the "Seq" tab. Copy the "Sequence with flank" and add the sequence to a fasta file with a defline ">RFTF:0000001655.001" id or if you want something simple like ">seq1", ">seq2", .... Repeat this process for the first 10 or so twist sites.
Now that you have a fasta file with binding sites and flanking regions, run MEME with the following command:
meme tfbs.fasta -dna -mod oops
You can scp your output directory "meme_out" to your computer with WinSCP or with scp. Investigate the contents of the directory. In particular, you could open the meme.html with a web browser.
How does your motif compare to the twist motif given here: twist motif
Feeling adventurous? You could try running MEME on the enhancer.fasta that you created for Project 3.a.
Copy and paste the following hairpin sequence to a fasta file called "hairpin.fasta":
>hairpin
ACAUACUUCUUUAUAUCCAUAUGAACCUGCUAAGCUAUGGAUGUAAAGAAGUAUGU
Fold this hairpin with RNAfold:
cat hairpin.fasta | RNAfold
Comment on the hairpin structure and the energy (given in parentheses). How big is the hairpin loop in terms of number of nucleotides?
Now use RNAduplex to compute the energy of the RNA duplex. Create a fasta file with the following arms of the hairpin:
>five
ACAUACUUCUUUAUAUCCAUA
>three
UAUGGAUGUAAAGAAGUAUGU
Now compute the duplex energy:
cat arms.fasta | RNAduplex
How much destabilizing energy does the hairping loop give? Try increasing the length of the hairpin loop by inserting nucleotides to the hairpin sequence above in increments of two. Note: You should choose nucleotides that aren't self-complementary to avoid creating more base pairs. Make a table with the difference in the hairpin energies and the duplex energies for the different lengths. How does the difference in energy change with the length of the loop? How does this compare to the equation given in class?
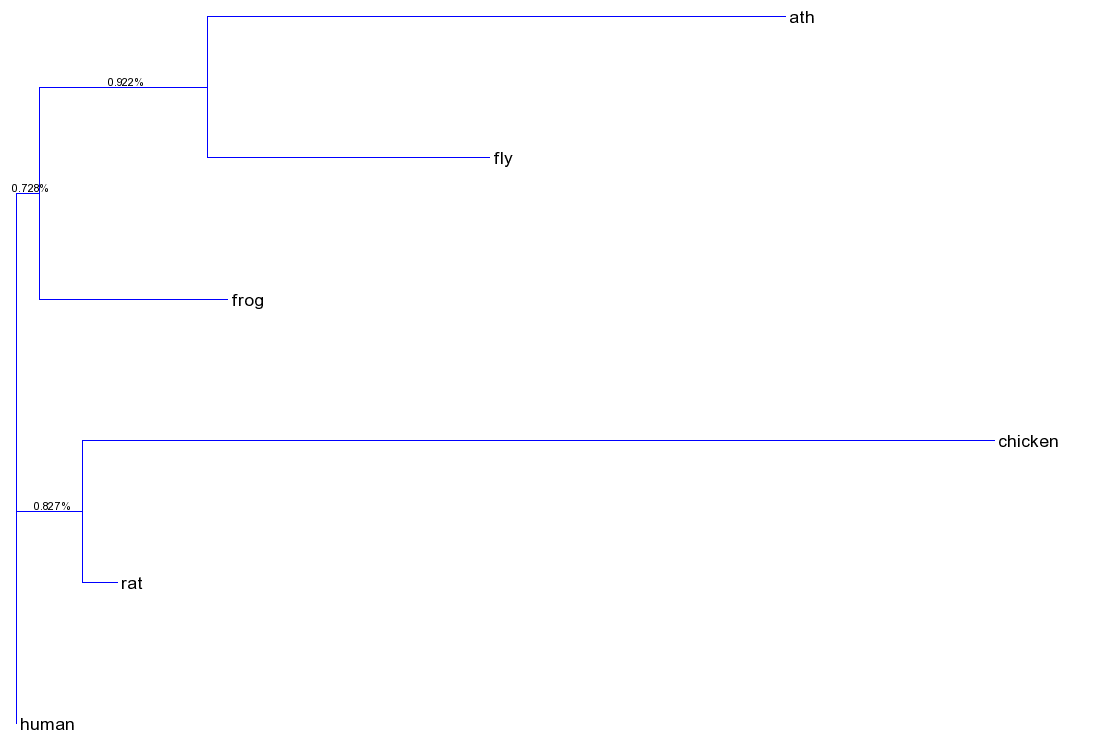
Download sequences in fasta format for a gene of interest from NCBI nucleotide (NCBI nucleotide) I downloaded 5S rRNA for Human (Homo sapiens), Mouse (mus musculus), Rat (Rattus norvegicus), Frog (Xenopus laevis), Chicken (Gallus gallus), Fly (Drosophila melanogaster) and Arabidopsis (A. thaliana). Build a fasta file containing each sequence and a defline. I shortened the defline for each species to make it easier to read later.
Create a multiple sequence alignment using ClustalW2. I pasted the fasta file into this web program here: ClustalW. Make sure you select DNA from the menu.
Once you have the alignment, convert your alignment to Plylip format. You can do this with the webtool here: ClustalW to Phylip. Download the Phylip formatted file.
Now build a phylogenetic tree using Phylip. Make sure you select DNA. You can experiment with different models such as JC69 or HKY85. How do the different models compare in the tree output? You can build the tree here: Phylip Tree builder
Once you have downloaded your output tree, check out the tree file. This is in Newick Tree format, which is a simple text file with the branch lengths for each branch of the resulting tree. You can visualize the tree using T.Rex, by pasting it here: T Rex. Download the tree, save it by naming it after the model you used (e.g. JC69) from Phylip.
Compare the trees for different genes, different types of molecules (mRNA vs rRNA for example) and for different models. Have fun!
Collect a set of homologous protein coding genes from NCBI Protein. Run MEME to find motifs. Next, run interproscan to identify domains. How do the domains and motifs compare?
Download the annotations for dm2 with wget here: flybase gene models gff
Download the Pol-II ChIP-chip data for the different 0-2hour Drosophila germ layers:
Pol-II ChIP-chip for Pipe mutants, Dorsal Ectoderm
Pol-II ChIP-chip for Rm9/10 mutants, Neuroectoderm
Pol-II ChIP-chip data for Toll-10b mutants, Mesoderm.
Download the script to compute the stalled genes here: computePolIIChIPChipStallingIndices.pl
You can run the script with the command:
perl computePolIIChIPChipStallingIndices.pl gene-models-gff Pol-II-fold-change-gff SI-threshold
For example:
perl computePolIIChIPChipStallingIndices.pl flyBaseGene.gff Pol2ChIPchip_Pipe.gff 4 > Pipe_geneIDList.txt
Now that you have a list of genes, you can transfer them to your computer. Head to GOEAST web tool here: GOEAST. Select Drosophila melanogaster from the species pull-down menu. I think you also need to select the Gene Symbol check box. The Step Optional button opens up numerous options. Let's use Hochberg (Benjamini-Hochberg) with an FDR of 0.05.
How do these GO Terms compare to the categories presented in lecture on Monday?
Bonus Question: Look for motifs in the forward strand for a selection of these top stalled genes.
The data for this project was taken from this paper: Wu et al.
Please download the data in the directory here: Yeast Data
What this data contains is 4 RNA-Seq files (reads from just chromosome VII), a tar file containing a bowtie index for the latest assembly of yeast, the genome sequence, and an annotation gtf file.
Step 1: Align the sequences with tophat. Please read more information on the options of tophat, check here: Tophat Manual
tophat -o sce_1n_R1_VII_sce_R64_1_1_tophat --no-coverage-search -no-novel-juncs -G sce_R64_1_1_ENSEMBL.gtf sce_R64_1_1 sce_1n_R1_VII.fastq
tophat -o sce_1n_R2_VII_sce_R64_1_1_tophat --no-coverage-search -no-novel-juncs -G sce_R64_1_1_ENSEMBL.gtf sce_R64_1_1 sce_1n_R2_VII.fastq
tophat -o sce_4n_R1_VII_sce_R64_1_1_tophat --no-coverage-search -no-novel-juncs -G sce_R64_1_1_ENSEMBL.gtf sce_R64_1_1 sce_4n_R1_VII.fastq
tophat -o sce_4n_R2_VII_sce_R64_1_1_tophat --no-coverage-search -no-novel-juncs -G sce_R64_1_1_ENSEMBL.gtf sce_R64_1_1 sce_4n_R2_VII.fastq
Step 2: Identify differentially expressed genes with Cuffdiff. Please read more information on the options of cuffdiffs, check here: Cuffdiff Manual
cuffdiff -L 4n,1n sce_R64_1_1_ENSEMBL.gtf sce_4n_R1_VII_sce_R64_1_1_tophat/accepted_hits.bam,sce_4n_R2_VII_sce_R64_1_1_tophat/accepted_hits.bam sce_1n_R1_VII_sce_R64_1_1_tophat/accepted_hits.bam,sce_1n_R2_VII_sce_R64_1_1_tophat/accepted_hits.bam
Do you find any differentially expressed genes? These will correspond to those significant at an FDR of 0.05, by default. There should be 4 genes. Are these 4 genes associated with any GO terms?
Bonus: If you want to take it further, you could re-do this project with the full dataset here: GEO link, although you should expect a decent runtime to do the full set.
Note: Some of the steps of this project take a long time, but usually not longer than 15-20 min at most, so plan accordingly.
The data for this project was taken from this paper: Integrated approaches reveal determinants of genome-wide binding and function of the transcription factor Pho4, Zhou et al. 2011. Please take a look at this paper. We will be looking at the data corresponding to Pho4 under the No Pi conditions.
This paper is studying the binding of the transcription factor Pho4, which binds to a sequence-specific motif described in the paper.
The raw data for this project is available at this GEO entry: GSE29506, but I have prepared the fastq files for download below.
Step 1: download the data and script from this directory with wget: Pho4 ChIP-Seq data and scripts. You will find 2 fastq files, and a perl script. The other data you will use was downloaded in Project 8. It would be a good idea to check the file sizes with "ls -l" to confirm that the download was complete.
$ ls -l *fastq -rw-r--r-- 1 dhendrix dhendrix 2817509732 Mar 10 23:23 sce_INPUT_NoPi_ChIPSeq.fastq -rw-r--r-- 1 dhendrix dhendrix 746715226 Mar 10 23:31 sce_Pho4_NoPi_ChIPSeq.fastq
Step 2: Align the fastq files to the genome: You need to align both the ChIP-Seq data and the INPUT (control) data.
bowtie2 -x sce_R64_1_1 -U sce_Pho4_NoPi_ChIPSeq.fastq -S sce_Pho4_NoPi_ChIPSeq.sam >& bwt1
bowtie2 -x sce_R64_1_1 -U sce_INPUT_NoPi_ChIPSeq.fastq -S sce_INPUT_NoPi_ChIPSeq.sam >& bwt2
Step 3: Convert the sam files to bed files for macs. You could also convert to bam or other formats, but this is one way to do it.
sam2bed sce_Pho4_NoPi_ChIPSeq.sam sce_Pho4_NoPi_ChIPSeq.bed
sam2bed sce_INPUT_NoPi_ChIPSeq.sam sce_INPUT_NoPi_ChIPSeq.bed
Step 4: Find the ChIP-Seq peaks with macs14. Note that we are using a stringent p-value threshold here to pull out only the strongest peaks.
macs14 -t sce_Pho4_NoPi_ChIPSeq.bed -c sce_INPUT_NoPi_ChIPSeq.bed -n Pho4_vs_INPUT -g 1.2e7 -f BED -p 1e-10 >& macs14.err
Macs will produce both peak bed files, and summit bed files. As you may have guessed, the peaks are the full enriched regions, and the summits are the tip of the peaks, and only constitute a small window of 2bp.
Step 5: Use the bed file of the summits to create a fasta file of of +/- 30bp around the summits:
perl ~/Scripts/bedToFasta.pl Pho4_vs_INPUT_summits.bed sce_R64_1_1.fa 30
This is the perl script you've downloaded above. Note that in general, this script is run with the inputs as follows:
perl bedToFasta.pl [bed file] [genome fasta] [sequence buffer]
Step 6: Run MEME to find motifs in the peak region:
meme Pho4_vs_INPUT_summits.fasta -mod zoops -dna -nmotifs 3 -maxw 8 -revcomp
If you did all the steps correctly, the binding site for Pho4 should be one of the motifs in the MEME output.
Questions to answer for this project: How many peaks do you find at this threshold? You can use the command "wc -l bedfile" to count the number of lines in a bed file produced for the peaks. Which motif corresponds to the motif in the paper? Include the LOGO for the motif you found in your results. How many instances of the motif did you find? What is the information content of the motif? Even though this was run with "zoops", you might still expect every peak to have an instance of the motif. How many of the peaks have an instance of the motif?